New Nature Communications Publication by Mann & Theis Groups Harnesses the Benefits of Large-scale Peptide Collisional Cross Section (CCS) Measurements and Deep Learning for 4D Proteomics
Bruker Corporation (Nasdaq: BRKR) today announces a seminal publication from the groups of Professors Matthias Mann and Fabian Theis in the journal Nature Communications with the title ‘Deep learning the collisional cross sections of the peptide universe from a million experimental values’ by Florian Meier et al. (doi.org/10.1038/s41467-021-21352-8)1.
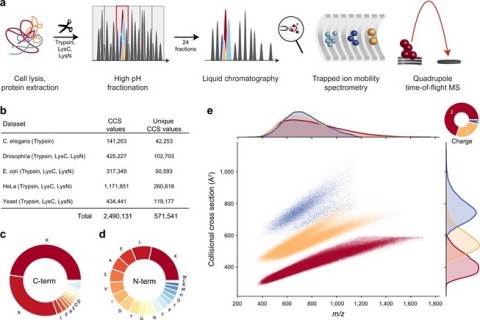
Fig. 1: Large-scale peptide collisional cross section (CCS) measurement with TIMS and PASEF. From "Deep learning the collisional cross sections of the peptide universe from a million experimental values". (a) Workflow from extraction of whole-cell proteomes through digestion, fractionation, and chromatographic separation of each fraction. The TIMS-quadrupole TOF mass spectrometer was operated in PASEF mode. (b) Overview of the CCS dataset in this study by organism. (c) Frequency of peptide C-terminal amino acids. (d) Frequency of peptide N-terminal amino acids. (e) Distribution of 559,979 unique data points, including modified sequence and charge state, in the CCS vs. m/z space color-coded by charge state. Density distributions for m/z and CCS are projected on the top and right axes, respectively. Source data are provided as a Source Data file. (Graphic: Business Wire)
The Nature Communications paper describes CCS values measured on the timsTOF pro as an essentially intrinsic property of the peptide ions, which can be used to improve confidence in peptide and protein group identification in 4D shotgun proteomics. Since mass spectrometry-based proteomics relies on accurate matching of acquired spectra against a database of protein sequences, accurate CCS values offer the benefit of narrowing down the list of candidates. This is essential for high sensitivity proteomics where low levels of peptide signals need to be accurately measured in a complex mixture, e.g. in plasma proteomics, peptidomics, immunopeptidomics or metaproteomics.
The publication summarizes a collaborative research effort led by Professor Matthias Mann, who holds dual appointments at the Max Planck Institute of Biochemistry in Martinsried, Germany and the Novo Nordisk Foundation Center for Protein Research at the University of Copenhagen in Denmark, together with the group of Professor Fabian Theis, who also holds dual appointments at the Helmholtz Center Munich in the German Research Center for Environmental Health, and in the Department of Mathematics at TU Munich, in Germany.
Lead author Dr. Florian Meier, now an Assistant Professor in Functional Proteomics at the Jena University Hospital in Germany, said: “The scale and precision of peptide CCS values in our data from the timsTOF pro was sufficient to train our deep learning model to accurately predict CCS values based only on the peptide sequence. This connection between the amino acids contained within a peptide sequence and its measured CCS has tremendous potential to increase the confidence of protein identification. Since the peptide CCS values are entirely determined by their linear amino acid sequences, they should be predictable with high accuracy and our deep learning model accurately predicted CCS values even for previously unobserved peptides. We acquired data from whole-proteome digests of five organisms, which resulted in the measurement of over two million CCS values, including about 500,000 unique peptides, making it by far the most comprehensive CCS data set to date.”
Professor Matthias Mann added: “The source code is publicly available so that further developments can be accelerated for training and prediction models of the human peptide universe. Conceptually, our CCS model could make dia-PASEF faster and less expensive by reducing the effort to generate libraries. Additionally, predicted CCS values should allow for the use of community libraries, such as the Pan Human library, a repository of over 10,000 human proteins, for targeted proteomics.”
Professor Fabian Theis stated: “Deep learning, in particular the used recurrent neural networks need a lot of samples to be predictive, so I was very happy when Matthias approached me and we jointly were able to predict and interpolate biochemical properties of peptides based only on their sequence. I personally liked the fact that we could thus impute CCS values also for many never before measured peptides."
Dr. Gary Kruppa, the Bruker Vice President for Proteomics, commented: “This paper showcases the tremendous potential of accurate CCS values for TIMS-PASEF methods in unbiased, deep 4D proteomics. The proven robustness, higher throughput and ultra-high sensitivity of the timsTOF platform is highly suitable for translational research. Large-scale peptide CCS values provide a fundamental advantage in the confidence of protein identification and quantitation in biomarker research in large cohort studies. Furthermore, the benefits of CCS values for improving confidence of identification are also applicable to other multiomics timsTOF workflows, such as metabolomics, lipidomics and glycomics. These are exciting times for our rapidly growing timsTOF user community.”
About Bruker Corporation (Nasdaq: BRKR)
Bruker is enabling scientists to make breakthrough discoveries and develop new applications that improve the quality of human life. Bruker’s high performance scientific instruments and high value analytical and diagnostic solutions enable scientists to explore life and materials at molecular, cellular and microscopic levels. In close cooperation with our customers, Bruker is enabling innovation, improved productivity and customer success in life science molecular and cell biology research, in applied and pharma applications, in microscopy and nanoanalysis, as well as in industrial applications. Bruker offers differentiated, high-value life science and diagnostics systems and solutions in preclinical imaging, clinical phenomics research, proteomics and multiomics, spatial and single-cell biology, functional structural and condensate biology, as well as in clinical microbiology and molecular diagnostics. For more information, please visit: www.bruker.com.
1 Meier, F., Köhler, N.D., Brunner, AD. et al. Deep learning the collisional cross sections of the peptide universe from a million experimental values. Nat Commun 12, 1185 (2021). https://doi.org/10.1038/s41467-021-21352-8
View source version on businesswire.com: https://www.businesswire.com/news/home/20210225005647/en/







